Insider Brief
- Researchers from Zhejiang University and HKUST (Guangzhou) have developed ProtET, an AI model that enables controllable protein editing through text-based instructions, improving functional protein design across multiple applications.
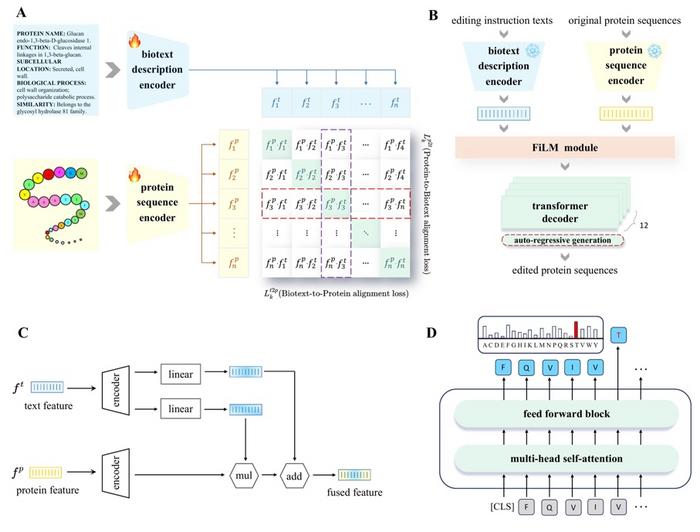
- Using a transformer-based architecture and contrastive learning, ProtET aligns protein sequences with natural language descriptions, demonstrating improvements in enzyme activity, stability, and antibody binding.
- Trained on over 67 million protein–biotext pairs, ProtET successfully designed SARS-CoV antibodies and optimized protein structures, highlighting its potential for biomedical research and synthetic biology.
- Image: The workflow and framework details of ProtET (Mingze Yin et al, published in Health Data Science)
PRESS RELEASE — Researchers from Zhejiang University and HKUST (Guangzhou) have developed a cutting-edge AI model, ProtET, that leverages multi-modal learning to enable controllable protein editing through text-based instructions. This innovative approach, published in Health Data Science, bridges the gap between biological language and protein sequence manipulation, enhancing functional protein design across domains like enzyme activity, stability, and antibody binding.
Proteins are the cornerstone of biological functions, and their precise modification holds immense potential for medical therapies, synthetic biology, and biotechnology. While traditional protein editing methods rely on labor-intensive laboratory experiments and single-task optimization models, ProtET introduces a transformer-structured encoder architecture and a hierarchical training paradigm. This model aligns protein sequences with natural language descriptions using contrastive learning, enabling intuitive, text-guided protein modifications.
The research team, led by Mingze Yin from Zhejiang University and Jintai Chen from HKUST (Guangzhou), trained ProtET on a dataset of over 67 million protein–biotext pairs, extracted from Swiss-Prot and TrEMBL databases. The model demonstrated exceptional performance across key benchmarks, improving protein stability by up to 16.9% and optimizing catalytic activities and antibody-specific binding.
“ProtET introduces a flexible, controllable approach to protein editing, allowing researchers to fine-tune biological functions with unparalleled precision,” said Mingze Yin, the study’s lead author.
The model successfully optimized protein sequences across different experimental scenarios, including enzyme catalytic activity, protein stability, and antibody-antigen interaction binding. In zero-shot tasks, ProtET designed SARS-CoV antibodies that formed stable and functional 3D structures, demonstrating its real-world applicability in biomedical research.
Looking ahead, the team envisions ProtET becoming a standard tool in protein engineering, paving the way for breakthroughs in synthetic biology, genetic therapies, and biopharmaceutical manufacturing.
This study marks a transformative step in AI-driven protein design, showcasing how cross-modal integration can unlock new horizons in scientific discovery and innovation.