Insider Brief
- DeepMind introduced AlphaProteo, an AI system that designs novel proteins capable of binding to specific target molecules, advancing drug discovery, disease research, and biosensor development.
- The system demonstrates 3- to 300-fold improvements in binding affinities compared to existing methods and has successfully created binders for critical proteins like VEGF-A and SARS-CoV-2 spike proteins.
- Despite its successes, AlphaProteo faced challenges in designing binders for TNFα, which the team says means further refinement is needed to tackle more complex biological targets.
DeepMind recently unveiled AlphaProteo, an AI-based system that generates novel protein binders tailored for specific molecular targets, according to a blog post and white paper.
The team is calling this a pivotal step in computational biology, with far-reaching implications for drug discovery, disease research and biosensor development.
Unlike AlphaFold, which predicts the structure of existing proteins, the team writes that AlphaProteo is “our first AI system for designing novel, high-strength protein binders to serve as building blocks for biological and health research. This technology has the potential to accelerate our understanding of biological processes, and aid the discovery of new drugs, the development of biosensors and more.”
This leap forward also holds the potential to accelerate key scientific fields by offering more precise and targeted solutions, the team suggests.
In the post, DeepMind emphasizes the crucial role proteins play in biology: “Every biological process in the body, from cell growth to immune responses, depends on interactions between molecules called proteins. Like a key to a lock, one protein can bind to another, helping regulate critical cellular processes.”
The challenge, however, lies in creating new proteins that can manipulate these interactions in meaningful ways. Traditional methods of designing protein binders are “time-intensive, requiring multiple rounds of extensive lab work” and often yield suboptimal results.
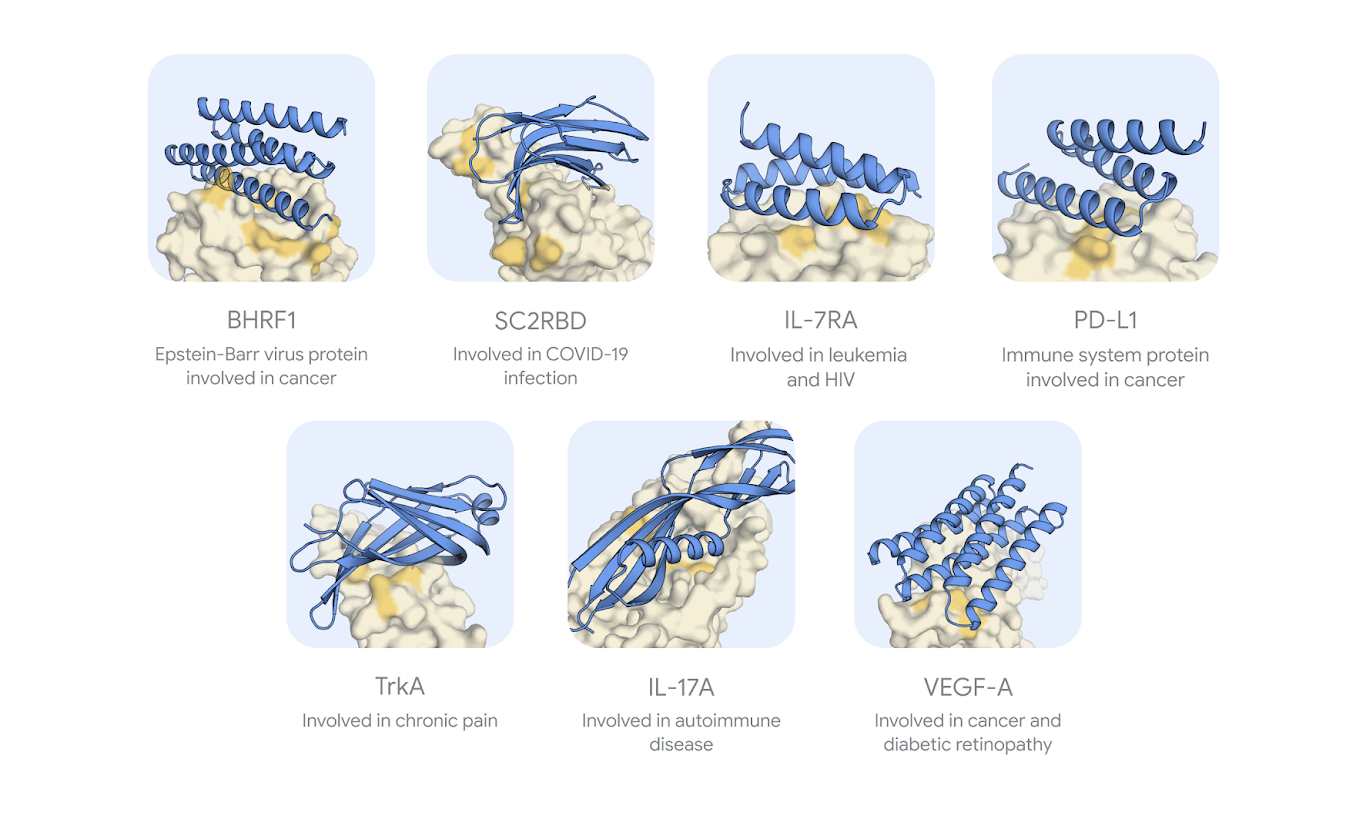
AlphaProteo promises to significantly streamline this process. It has been tested on multiple target proteins, including vascular endothelial growth factor, or VEGF-A. The team points out that the protein plays a critical role in the formation of new blood vessels, a process known as angiogenesis. This is particularly important in both normal bodily functions, like wound healing, and in pathological conditions, such as cancer.
In tumors, VEGF stimulates the growth of new blood vessels that supply nutrients to cancer cells, aiding their proliferation. Because of its role in promoting tumor growth, VEGF has become a key target in cancer therapies, with drugs designed to inhibit its function in order to restrict blood supply to tumors, slowing their progression. VEGF is also implicated in diabetic retinopathy and age-related macular degeneration, making it a target for treatments in various medical fields., a molecule associated with cancer and diabetic complications.
DeepMind reports that “this is the first time an AI tool has been able to design a successful protein binder for VEGF-A”. Designing this binder could inform future work to create innovative therapeutic strategies in oncology and other fields.
AlphaProteo was also tested on the SARS-CoV-2 spike protein receptor-binding domain (SC2RBD), the key protein involved in COVID-19 infection.
In collaboration with the Francis Crick Institute, DeepMind confirmed that the protein binders developed by AlphaProteo could serve as a weapon against Covid.
The write: “The research groups confirmed that the binding interactions of these binders were indeed similar to what AlphaProteo had predicted. Additionally, the groups confirmed that the binders have useful biological function. For example, some of our SC2RBD binders were shown to prevent SARS-CoV-2 and some of its variants from infecting cells.”
This suggests that AlphaProteo could be used to develop antiviral therapies for current and future pandemics.
While AlphaProteo has shown tremendous promise, it also has limitations.
When tested against TNFα, a protein involved in autoimmune diseases, the system was unable to produce an effective binder. DeepMind candidly acknowledges this shortfall, stating:
“We selected TNFɑ to robustly challenge AlphaProteo, as computational analysis showed that it would be extremely difficult to design binders against. We will continue to improve and expand AlphaProteo’s capabilities with the goal of eventually addressing such challenging targets.”
This demonstrates that, although AlphaProteo represents a significant advancement, there is still work to be done in refining the technology for more complex targets.
They add: “Achieving strong binding is usually only the first step in designing proteins that might be useful for practical applications, and there are many more bioengineering obstacles to overcome in the research and development process.”
Experimental Success
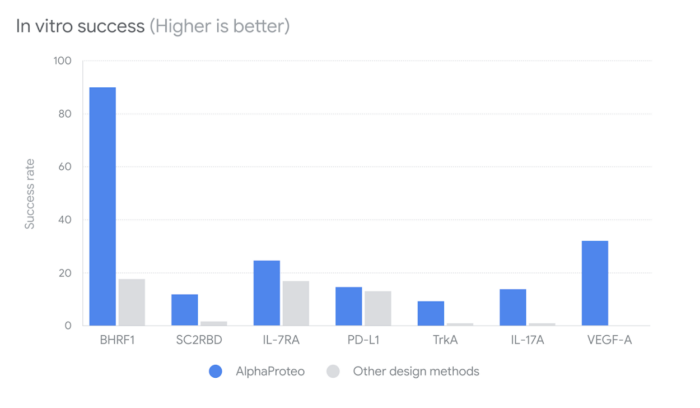
In terms of experimental results, AlphaProteo’s success rates are impressive, the research paper reported: “For seven of the targets, between 9% and 88% of the designs tested in the wet lab were experimentally verified as successful binders. These figures are higher than the best existing method and 5- to 100-fold higher than other methods. For one of these targets we report the first computationally designed binders.”
Additionally, the team detailed a jump in binding affinities: “We obtain binders with 80-960 picomolar affinities to four targets and low-nanomolar affinities to another three without needing high-throughput screening or experimental affinity optimization. For the seven targets, our designs have 3- to 300-fold better binding affinities than the best previous designed binder.”
Validation of these designed proteins was carried out using advanced techniques such as cryo-electron microscopy (cryo-EM) and X-ray crystallography, with DeepMind confirming that these methods helped confirm the designed binder and binder-target complex structures.
AlphaProteo is poised to revolutionize various fields beyond just healthcare. According to DeepMind: Protein design is a fast-evolving technology that holds lots of potential for advancing science in everything from understanding the factors that cause disease, to accelerating diagnostic test development for virus outbreaks, supporting more sustainable manufacturing processes, and even cleaning contaminants from the environment.
Responsible Development
DeepMind is committed to ensuring that AlphaProteo is developed responsibly, according to the blog post. The team has integrated biosecurity measures into their research, noting that they are “working with leading external experts to inform our phased approach to sharing this work” to address any potential risks.
Looking ahead, DeepMind aims to collaborate with the broader scientific community to further explore AlphaProteo’s potential.
They add: “Going forward, we’ll be working with the scientific community to leverage AlphaProteo on impactful biology problems and understand its limitations. We’ve also been exploring its drug design applications at Isomorphic Labs, and are excited for what the future holds. At the same time, we’re continuing to improve the success rate and affinity of AlphaProteo’s algorithms, expanding the range of design problems it can tackle, and working with researchers in machine learning, structural biology, biochemistry and other disciplines to develop a responsible and more comprehensive protein design offering for the community.”
The collaboration included researchers from Google DeepMind, The Chromatin Structure and Mobile DNA Laboratory at The Francis Crick Institute in London, the COVID Surveillance Unit at The Francis Crick Institute in London, the Cellular Adaptive Behaviour Laboratory at The Francis Crick Institute in London, the Department of Informatics at King’s College London, and the RNA Virus Replication Laboratory at The Francis Crick Institute in London.