Insider Brief
- Researchers at Pusan National University developed MoGLo-Net, a deep learning model that reconstructs 3D images from 2D photoacoustic and ultrasound scans without relying on external sensors, enhancing accuracy and reducing equipment complexity.
- MoGLo-Net uses tissue speckle patterns and combines a ResNet-driven encoder with an LSTM-based motion estimator to track transducer motion and generate 3D images, outperforming state-of-the-art models in tests across multiple datasets.
- Published in IEEE Transactions on Medical Imaging, this advancement promises safer, more effective diagnostic imaging and interventions by expanding access to affordable 3D ultrasound technology in clinical settings.
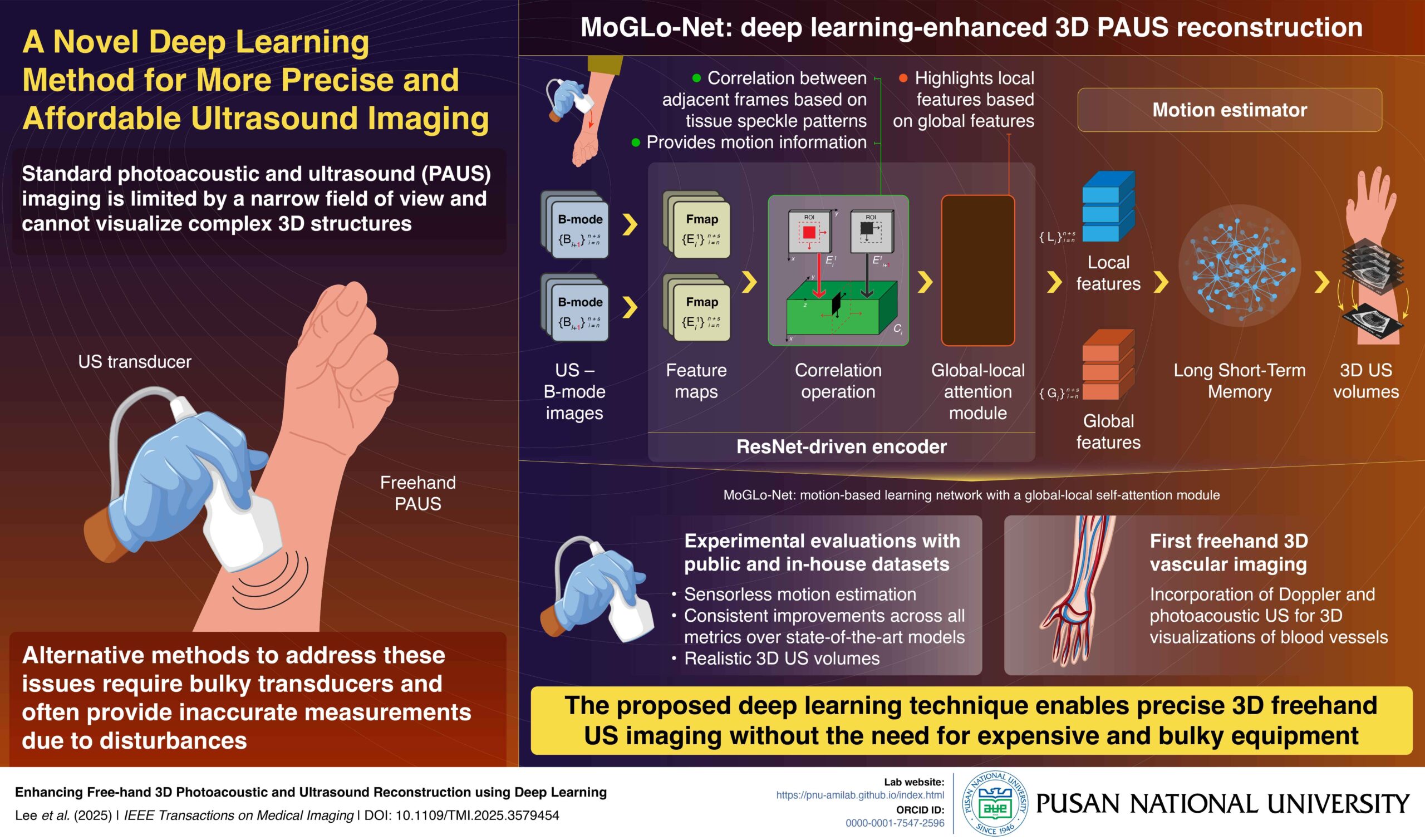
PRESS RELEASE – Conventional handheld photoacoustic and ultrasound Imaging (PAUS), while offering flexibility, offers only a narrow view of the target region, providing limited information on its structure. Alternative methods require external sensors and bulky equipment, which can also make measurements inaccurate. In a new study, researchers developed a MoGLo-Net, a deep-learning model for 3D reconstruction of 2D PAUS images. This method does not require any external sensors and can make treatments more accessible, safer, and effective.
Ultrasound (US) imaging is a widely employed diagnostic tool used for real-time imaging of various organs and tissues using ultrasonic sound waves. The waves are sent into the body, and images are created based on how the waves reflect off internal tissues and organs. It is used for guiding many medical procedures, including biopsies and injections, and is important for dynamic monitoring of blood vessels. When the US is combined with photoacoustic (PA) imaging, where laser light pulses are used to produce sound waves in tissues, the resulting technique, called PAUS imaging, offers enhanced imaging capabilities.
In PAUS imaging, a doctor holds a transducer, responsible for emitting US or laser pulses, and guides it over the target region. While this configuration is flexible, it captures only a small two-dimensional (2D) area of the target, offering a limited understanding of its three-dimensional (3D) structure. Though some transducers offer complete 3D imaging, they are expensive and have a limited field of view. An alternative method is the 3D freehand method, in which 2D images scanned (obtained) by sweeping a transducer over the body surface are stitched together to create a 3D view. A key challenge in this technique, however, is the precise tracking of transducer motion, requiring expensive and bulky external sensors that often provide inaccurate measurements.
To address this issue, a research team from Korea, led by Associate Professor MinWoo Kim from the School of Biomedical Convergence Engineering and the Center for Artificial Intelligence Research at Pusan National University, developed a deep learning model called MoGLo-Net. “MoGLo-Net automatically tracks the motion of the ultrasound transducer without using any external sensors, by using tissue speckle data,” explains Prof. Kim. “This model can create clear 3D images from 2D ultrasound scans, helping doctors understand what’s happening inside the body more easily, and making better decisions for treatment.” Their study was published in the journal IEEE Transactions on Medical Imaging on 13 June, 2025.
MoGLo-Net estimates transducer motion directly from US B-mode image sequences. It consists of two main parts: an encoder driven by the ResNet deep learning framework, and a motion estimator, powered by the Long-Short Term Memory (LSTM) neural network. The ResNET-driven encoder consists of special blocks that can extract the correlation between consecutive images based on tissue speckle patterns, a technique known as correlation operation. This helps capture both in-plane and out-of-plane motion.
The information is fed into a novel self-attention mechanism in the encoder that highlights local features from specific regions in images, based on global features that summarize information from the entire image. The resulting final features are passed on to the LSTM-based motion estimator, which estimates the motion of the transducer over time, leveraging long-term memory. Furthermore, the model employs customized loss functions that ensure accuracy.
The researchers tested MoGLo-Net in diverse conditions using both proprietary and public datasets and found that it outperformed state-of-the-art models on all metrics, producing more realistic 3D US images. In a first for the field, the researchers also combined ultrasound and photoacoustic data to reconstruct 3D images of blood vessels using this model.
“Our model holds immense clinical potential in diagnostic imaging and related interventions,” remarks Prof. Kim. “By offering clear 3D images of various bodily structures, this technology can help make medical procedures safer and more effective. Importantly, by removing the need for bulky sensors, this technology democratizes the use of ultrasound, making it accessible to clinics where specialists may not be available.”
This innovation marks a major milestone in ultrasound imaging, paving the way for more accurate, efficient, and affordable healthcare for all.